Me to son: You have way too many toys! Go pick one and we’ll donate it to charity.
*10 minutes later*
Me: woah woah woah slow down, too many memories with that one.
19.01.2025 15:58 — 👍 2 🔁 0 💬 0 📌 0
I’d guess that a lot of ancestral exploration was driven toward some material gain. For example, “I wonder if there’s some food over there…”
14.01.2025 03:15 — 👍 0 🔁 0 💬 2 📌 0
My wife woke up angry because Ariana Grande stole me from her in a dream.
12.01.2025 19:00 — 👍 2 🔁 0 💬 0 📌 0
As kids, my mom used to regularly take me and my sister to public recycling dumpsters, have us climb in, and we’d dig around to find coupons in magazines and newspapers. We were not poor.
12.01.2025 18:59 — 👍 1 🔁 0 💬 0 📌 0
My grandfather compiled a history of all of my ancestors on his side going back 5 generations, 62 people in total, some born in the mid 1700s. I can confirm that life is much better now than it was then.
19.12.2024 15:36 — 👍 2 🔁 0 💬 0 📌 0
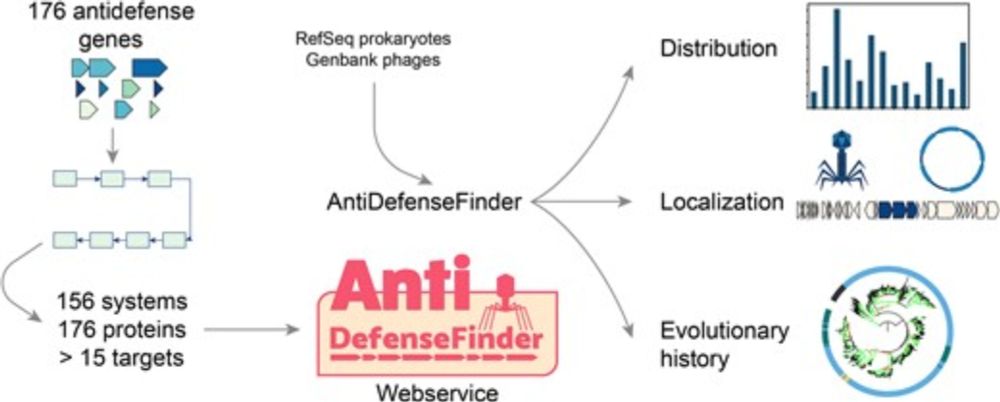
Exploring the diversity of anti-defense systems across prokaryotes, phages and mobile genetic elements
Abstract. The co-evolution of prokaryotes, phages and mobile genetic elements (MGEs) has driven the diversification of defense and anti-defense systems ali
Out in NAR, AntiDefenseFinder & our "systematic" analysis of inhibitors of antiphage systems wt Bondy-Denomy lab led by @ftesson.bsky.social and E. Huiting.
Not so "systematic" as very few known, fun stuff on MGE, antidefense islands and cool phages exaptations!
academic.oup.com/nar/advance-...
16.12.2024 08:53 — 👍 110 🔁 50 💬 0 📌 1
This whole story is amazing, intuitive in retrospect, and likely exceptionally important medically. Do read the paper and the whole thread!
11.12.2024 19:20 — 👍 13 🔁 2 💬 1 📌 0
Isn’t the solution to ChatGPT doing everyone’s homework to just have more exams and to weight them more heavily?
04.12.2024 03:02 — 👍 0 🔁 0 💬 0 📌 0
Come join us at @arcinstitute.org !
03.12.2024 00:54 — 👍 15 🔁 6 💬 0 📌 0
If the government is going to start banning food dyes and vaccines I only ask that they ban such things in a time-staggered way throughout the country so we can get some nice natural experiment data.
02.12.2024 03:57 — 👍 1 🔁 0 💬 0 📌 0
I hereby announce my candidacy for the presidency in 2028. If you vote for me, I will give you complete immunity for all crimes you commit between now and the end of my time in office.
02.12.2024 03:43 — 👍 1 🔁 0 💬 0 📌 0
Where can I get my hands on a woolly mammoth genome? Was expecting more than mitochondria on NCBI.
27.11.2024 00:15 — 👍 3 🔁 2 💬 1 📌 0
Yet more evidence that transfer learning of sequence-only PLMs does not benefit from scale beyond 650M params 🧵
25.11.2024 07:34 — 👍 78 🔁 20 💬 7 📌 3
Microbiology will always be one of the most satisfying fields of natural science. Nigh-infinite levels of biodiversity, perpetual room for novel findings, and an evergreen requirement for the advancement of human civilization. 10/10. Big fan. Would recommend.
🦠💪
25.11.2024 13:12 — 👍 315 🔁 36 💬 14 📌 2
That’s the last of my faculty applications! Now we wait🤞
25.11.2024 06:52 — 👍 1 🔁 0 💬 1 📌 0
Could really use the bookmark button….
24.11.2024 04:15 — 👍 0 🔁 0 💬 1 📌 0
Really is impressive to see the growth here!
24.11.2024 03:18 — 👍 2 🔁 0 💬 0 📌 0
Wow
24.11.2024 03:05 — 👍 1 🔁 0 💬 0 📌 0
Our Big Fantastic Virus Database (BFVD) is now published NAR! It contains protein structure predictions of major viral clades, enhanced by petabase-scale homology search and it's explorable on the web.
🌐 bfvd.foldseek.com
💾 bfvd.steineggerlab.workers.dev
📄 academic.oup.com/nar/advance-...
23.11.2024 21:12 — 👍 339 🔁 127 💬 6 📌 5
We've long been able to find new phages and figure out what they interact with. But what about the other direction: if you have a gene, like an antibiotic resistance efflux pump, or a phage defense system, and you want to find phages that do or do not interact with it?
Now you can!
23.11.2024 18:49 — 👍 83 🔁 35 💬 4 📌 0
Thanks for adding me! This is great.
20.11.2024 00:49 — 👍 1 🔁 0 💬 0 📌 0

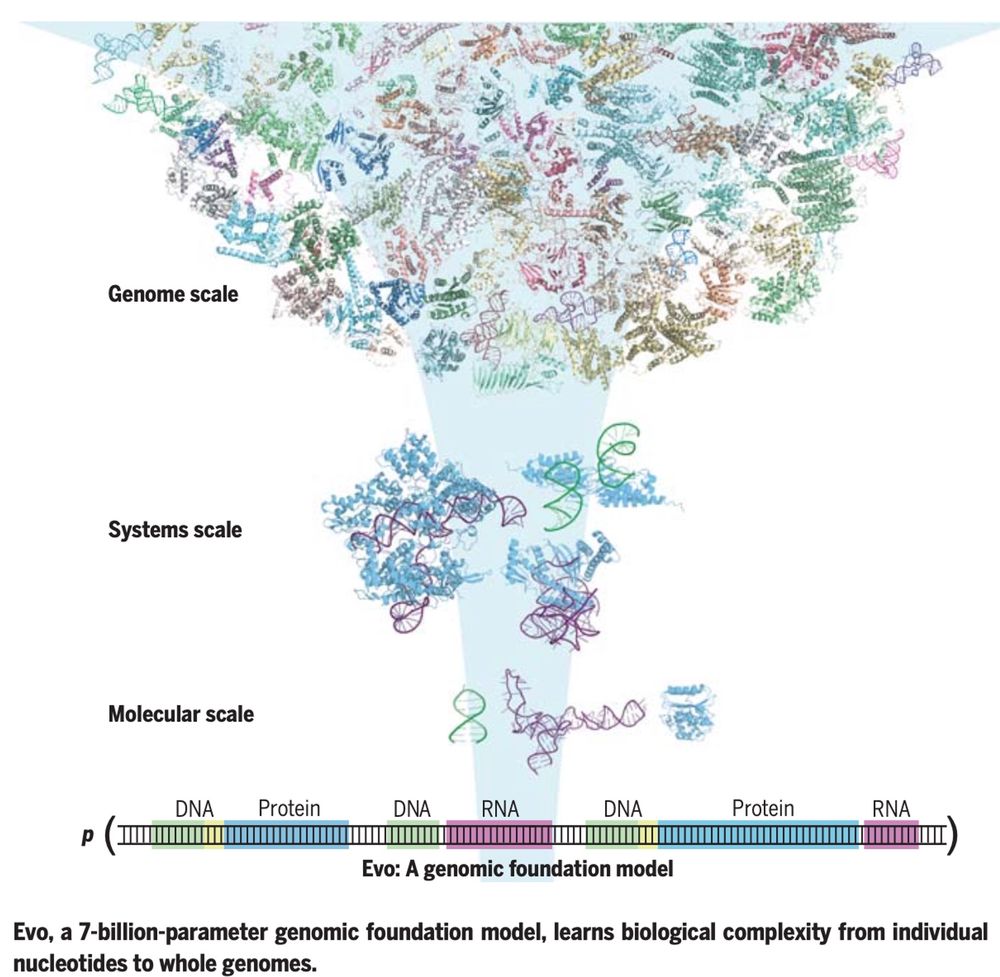
New at Science
Evo, a large language of life (LLLM) genomic foundation model, predicting & generating tasks from molecular to genomic scale
science.org/doi/10.1126/...
14.11.2024 19:16 — 👍 207 🔁 40 💬 3 📌 1
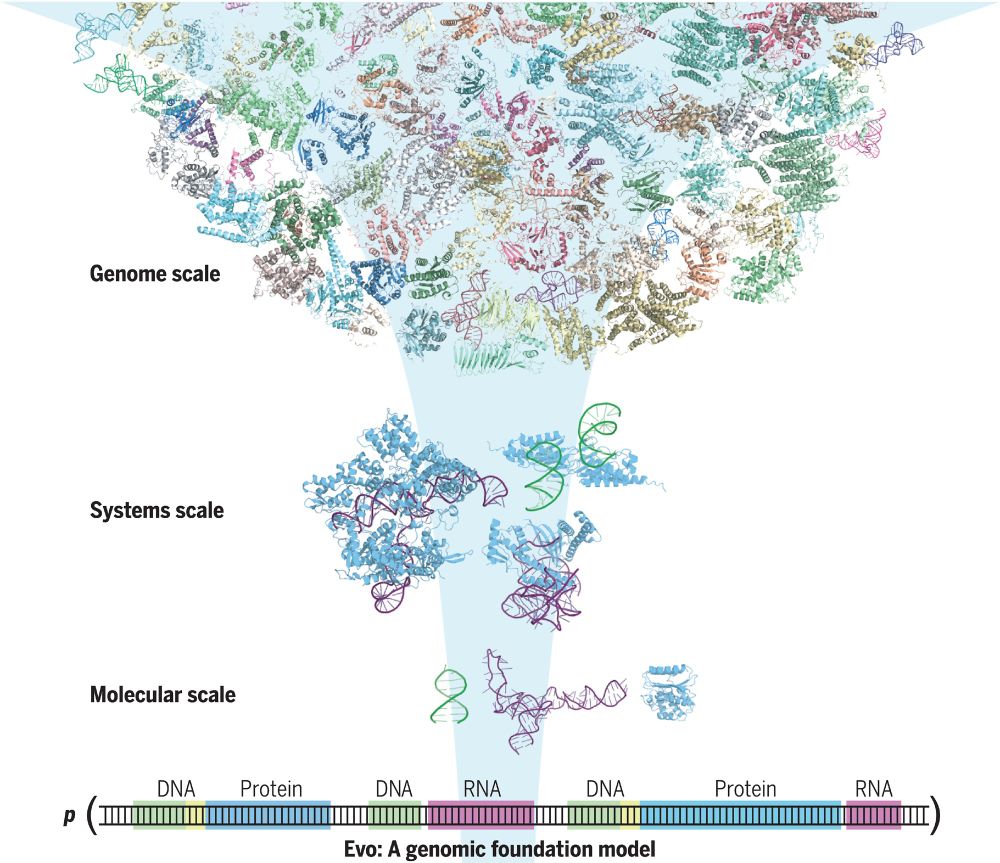
Evo, a 7-billion-parameter genomic foundation model, learns biological complexity from individual nucleotides to whole genomes.
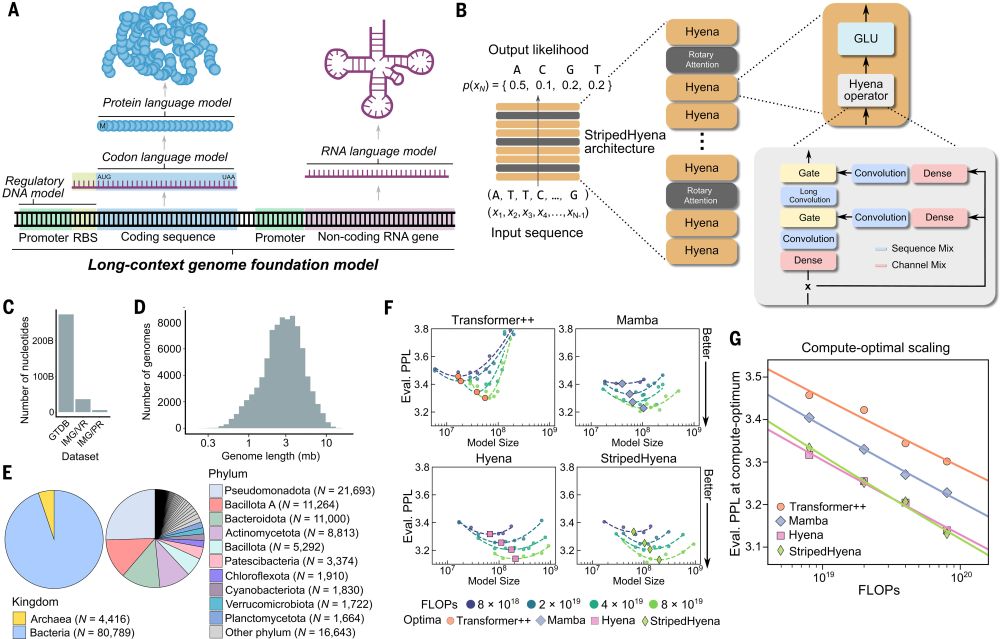
Pretraining a genomic foundation model across prokaryotic life.
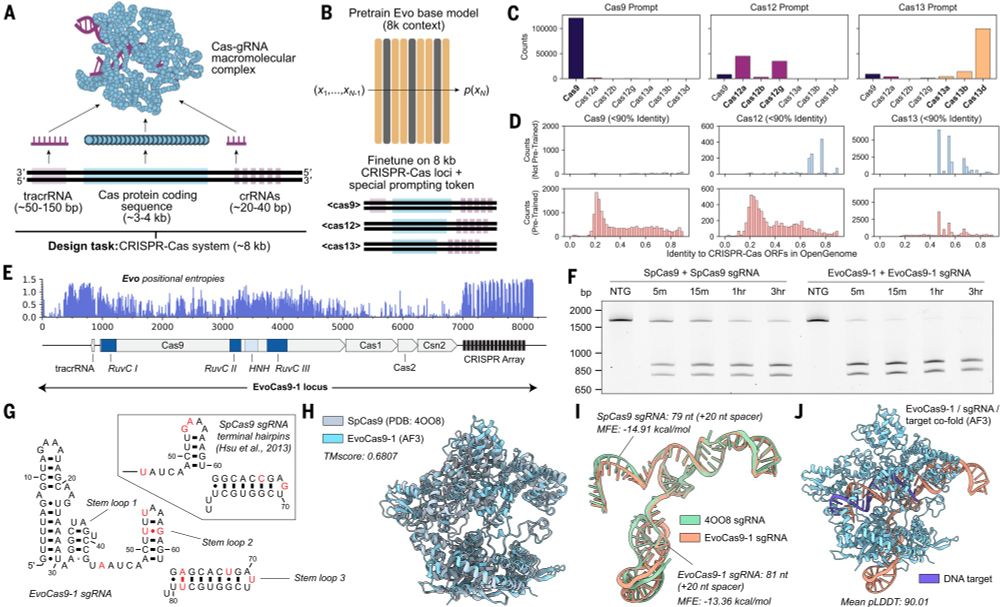
Fine-tuning on CRISPR-Cas sequences enables generative design of protein-RNA complexes.
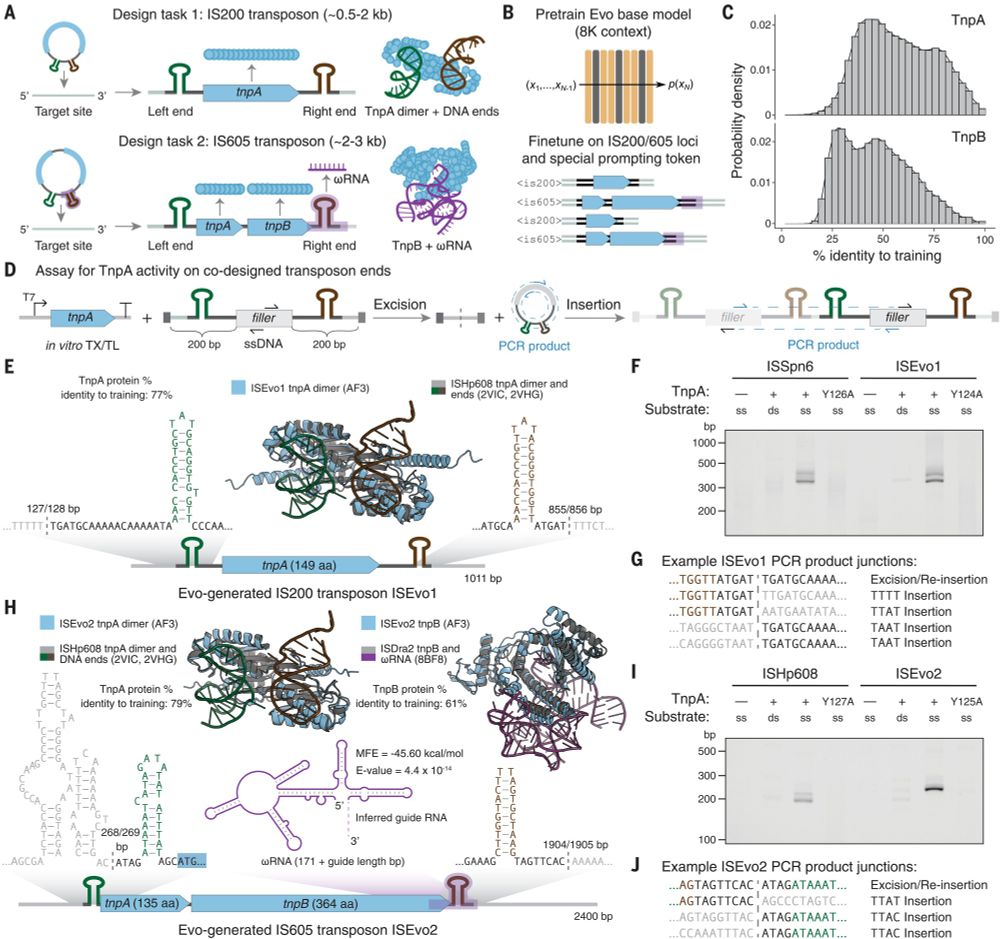
Fig. 4. Fine-tuning on IS200/IS605 sequences enables generative design of transposable biological systems.
Evo: A genomic language model of prokaryote genomes generates functional cas9 proteins and transposons.
@brianhiestand.bsky.social
www.science.org/doi/10.1126/...
14.11.2024 20:53 — 👍 59 🔁 27 💬 0 📌 1

Really cool model from Brian Hie and Patrick Hsu! www.science.org/doi/10.1126/...
14.11.2024 19:35 — 👍 18 🔁 8 💬 0 📌 0
The Great Kettlebell Swing Forward
14.11.2024 22:40 — 👍 0 🔁 0 💬 0 📌 0
Contents | Science 386, 6723
Thrilled to see this published! Lots of progress since our preprint, including experimental validation of Evo-generated Cas9 and IS200/IS605 transposases. I am definitely a true believer when it comes to DNA language models, they have quickly become central to my discovery efforts. bit.ly/3OsmUPr
14.11.2024 21:07 — 👍 5 🔁 2 💬 0 📌 0
Where is this tree from?
14.11.2024 00:03 — 👍 1 🔁 0 💬 2 📌 0
What are some unwritten rules in academia that would be helpful for new folks to know?
One rule I learned the hard way was that you don't reach out to other PIs about collaborating without discussing it with your PI first.
06.10.2023 18:35 — 👍 0 🔁 0 💬 0 📌 0
Does anyone know where I could find some measure of brain drain per year for each country?
04.10.2023 21:17 — 👍 0 🔁 0 💬 0 📌 0
Nature Portfolio’s high-quality products and services across the life, physical, chemical and applied sciences is dedicated to serving the scientific community.
I write software for Biologists. Founder @fulcrumgenomics, Bioinformatician, Computer Scientist, Coder, Father of 2xGirls. Opinions are my own.
Group Leader - Reader (Assoc. Professor) in AI and Cancer Epigenetics at Blizard Institute, Queen Mary University of London.
#GeneRegulation, #Chromatin, #Epigenetics, #AI, #bioinformatics
https://www.qmul.ac.uk/blizard/all-staff/profiles/radu-zabet.html
Group leader at MRC Human Genetics Unit, University of Edinburgh. Interdisciplinary research on disease #epigenetics. Part-time solo dad. Occasional music and climbing.
https://institute-genetics-cancer.ed.ac.uk/research/research-groups-a-z/sproul-group
Assistant Professor of Cancer Biology at Weill Cornell Medicine — NIH NCI R00 Funded — Chromatin & Epigenetics in Cancer and Immunity.
Group leader of the Stochastic Systems Biology Lab at IGBMC - CNRS - University of Strasbourg. Models of gene regulation based on biophysics-informed deep learning: https://www.igbmc.fr/molina
🔬 Cell adhesion networks lab
🥼 A new cell systems biology team
📍 University of Manchester, UK
🔗 https://linktr.ee/TheByronLab
https://www.helmholtz-munich.de/en/ife
Assistant Professor, Tish Cancer Institute @Mount Sinai. Colorectal Cancer. Oncofetal reprogramming. Views are my own. "By land and by sea, Carthage will rise again"
https://profiles.mountsinai.org/slim-mzoughi
Researcher in Epigenetics, CRISPRing around. MSCA Postdoctoral Fellow at EMBL Rome (Hackett lab). Before: Max Planck for Molecular Genetics (Ph.D.), Exeter Uni, Pasteur Institute, Sapienza Uni - Pessimismo dell' intelligenza/Ottimismo della volontà.
Lorry Lokey Chair & Professor of Bioengineering, Knight Campus, University of Oregon. evolution | ecology | genetics | genomics | evodevo | systems biology | epigenetics | host-microbe. Educational empowerment & increasing opportunities for all in science.
Post doc 🧬👩🏻🔬👩🏻💻 in @thecesarelab.bsky.social at @cmri.bsky.social | using insights from epigenetics and genome stability to improve pediatric cancer treatment | Optimist 😊 | Runner 🏃🏻♀️ | Wife & Mum 💛🌻
Developmental biologist working at CNRS/IGMM/University of Montpellier, interested by gene regulation in drosophila embryos. we use quantitative imaging to monitor mRNA lifecycle.
The Anja Groth group in Copenhagen studies chromatin replication, epigenetic memory and (epi)genome stability in the context of mitotic cell division
Group Leader at MPI Molgen, Berlin, Germany
Interested in quantitative biology, gene regulation, epigenetics, system biology and X-chromosome inactivation.
https://www.molgen.mpg.de/schulz-lab
Independent Group Leader at the MPI-IE passionate about mechanisms governing embryonic development with a focus on transposable elements🧬 Postdoc @MundlosLab 🇩🇪| PhD from @Bourchis lab 🇫🇷| She/her
https://www.julianeglaserlab.com/
A bit of 3D gene regulation, single-cell omics and transgenic models.
Born and raised in the bay of Algeciras. Enjoying Science and Flamenco at CABD, Seville.
Lab website: https://lupianezlab.github.io/Website/
Email: dario.lupianez@csic.es
Biologist studying transposons, gene regulation and chromatin in the Jullien group.
Postdoc @ IBMP (Strasbourg, France).
@ibmp-cnrs.bsky.social
Group leader, transcription dynamics in single cells, NKI
Associate Professor | UNC Chapel Hill, Pharmacology Department | Lineberger Comprehensive Cancer Center | Research in Endocrinology, Epigenetics, & Wnt signaling| Views my own. https://www.med.unc.edu/pharm/pruittlab/team/