Overview | 2025 NCI Spring School on Algorithmic Cancer Biology | NCIF Conferences
SSACB 2025 rescheduled to Aug 18-22! We are inviting students and postdocs broadly interested in computational cancer to 2025 NCI Summer School on Algorithmic Cancer Biology. Please apply and spread the word: ncifrederick.cancer.gov/events/confe...
20.06.2025 13:31 — 👍 0 🔁 0 💬 0 📌 0

The Department of Human Genetics at the University of Utah is sponsoring the Rising Stars in Genetics and Genomics symposium!
- We are seeking nominations bu June 1.
- September 18-19, 2025
- Please share with the star postdocs that you know.
docs.google.com/forms/d/e/1F...
28.04.2025 17:20 — 👍 53 🔁 43 💬 1 📌 1
🚨 Pre-print alert! 🚨
We generated chromosome-scale diploid genome assemblies for the widely used fibroblast lines BJ and IMR-90.
These models of replicative senescence and telomere shortening now have fully resolved telomeres, subtelomeres, and centromeres. 🧬
🧵👇
16.04.2025 18:05 — 👍 9 🔁 5 💬 1 📌 0

Our lab at the NIH (Bethesda, MD) is looking for postdoctoral fellows to join our team studying the molecular mechanisms of protein trafficking and their links to neurodevelopmental disorders starting on or after 10-1-2025. Send your application to juan.bonifacino@nih.gov. Please share and repost!
13.04.2025 16:36 — 👍 89 🔁 80 💬 5 📌 8
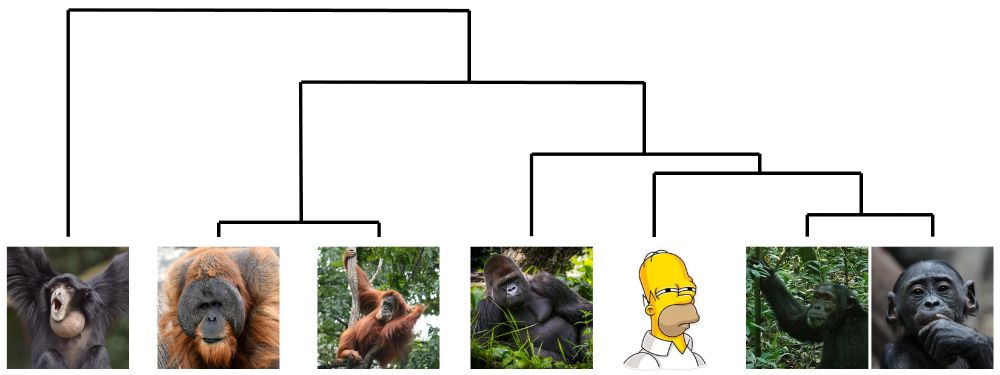
A phylogeny of the 7 ape genomes that have now been completed from "T2T", with Homer Simpson representing mankind.
A project five years in the making, we've now published complete "T2T" genomes for six additional ape species! It turns out that finishing (and analyzing) six genomes is slightly more work than one... doi.org/10.1038/s415...
09.04.2025 21:31 — 👍 156 🔁 77 💬 5 📌 3
A milestone for our lab! Here's a full access link: rdcu.be/egmYb
05.04.2025 16:40 — 👍 22 🔁 3 💬 1 📌 0

a picture of a pikachu with the words " i still have hope " written below it
ALT: a picture of a pikachu with the words " i still have hope " written below it
Some glimmers of hope (but mostly confusion) this morning.
I have heard that no intramural tenure track investigators were supposed to be on the termination list although some folks clearly got termination notices.
I am actively checking into this every way I can.
1/2
16.02.2025 12:44 — 👍 75 🔁 25 💬 4 📌 0

a man standing in front of a crowd with the words everybody panic
Alt: a man standing in front of a crowd with the words everybody panic
Short update on NIH
The situation is very confusing, a not just to me. Folks who were told the were to be terminated did not receive any communication for a day and a half, people who were “on the list” have not received anything as of yet. Some folks who had been terminated have be reinstated
1/2
16.02.2025 15:53 — 👍 125 🔁 31 💬 2 📌 3
Overview | 2025 NCI Spring School on Algorithmic Cancer Biology | NCIF Conferences
This is becoming a tradition! We are inviting students and postdocs broadly interested in computational cancer to 2025 NCI Spring School on Algorithmic Cancer Biology. Happening Mar 10-14. Please apply and spread the word: ncifrederick.cancer.gov/events/confe...
14.01.2025 22:41 — 👍 6 🔁 5 💬 0 📌 0

Biology of Genomes
Cold Spring Harbor Laboratory Meetings & Courses -- a private, non-profit institution with research programs in cancer, neuroscience, plant biology, genomics, bioinformatics.
Biology of Genomes 2025 is coming up:
- meeting dates: May 6 - 10, 2025
- abstract deadline: Feb 14, 2025
We hope to see you there for another great meeting with a fantastic speaker lineup!
meetings.cshl.edu/meetings.asp...
03.12.2024 19:21 — 👍 67 🔁 34 💬 1 📌 2

Genomics+Bioinformatics Starter Pack 🧬🖥️
Join the conversation
I created a genomics+bioinformatics starter pack. If I left you off, *please* reply and I'll add you! go.bsky.app/B5YYBfq
22.10.2024 12:34 — 👍 176 🔁 106 💬 99 📌 7
#Genetics Starter Pack ~100 accounts in genetics and genomics. Apologies to all the lovely people I missed.
go.bsky.app/JcbBNq5
08.11.2024 18:55 — 👍 102 🔁 79 💬 25 📌 5
I made a starter pack for algorithmic genomics. It's certainly incomplete, but already has a ton of awesome peeps. Let me know if you know people I should add (with a focus on algorithms and data structures in genomics)
go.bsky.app/TRWCnZs
12.11.2024 14:03 — 👍 141 🔁 65 💬 25 📌 2
I created a long-read sequencing starter pack. I'm still finding people on here, so apologies if I missed you. Please suggest people who should be added and I'll add them! go.bsky.app/JGkefsJ
11.11.2024 05:05 — 👍 38 🔁 23 💬 16 📌 1

Six weeks left until the abstract deadline for Biology of Genomes at CSHL #BoG24! We have a stellar lineup of keynotes and discussion leaders (=~invited speakers), and look forward to another brilliant meeting in May 7-11. Abstract submission & info:
meetings.cshl.edu/meetings.asp...
05.01.2024 13:22 — 👍 13 🔁 17 💬 2 📌 0
NCI Spring School on Algorithmic Cancer Biology is happening again Apr 1-5 2024! The registration is now open, we invite trainees broadly interested in computational cancer biology to participate. Please apply and spread the word! ncifrederick.cancer.gov/events/confe...
20.11.2023 21:48 — 👍 10 🔁 7 💬 0 📌 0

I am very excited about guest editing a special issue in Genome Research (@genomeresearch.bsky.social )
about long reads DNA Sequencing Applications with Ana Conesa & Alexander Hoischen. This is such a timely and fast moving topic about all its applications, innovations etc. bit.ly/grcallforpap...
06.11.2023 14:26 — 👍 14 🔁 10 💬 0 📌 0
GitHub - marbl/HG002: A complete diploid human genome
A complete diploid human genome. Contribute to marbl/HG002 development by creating an account on GitHub.
We are excited to release v1.0 of our HG002 genome benchmark today! Our goal with the Q100 project is to create a complete and perfectly accurate representation of the diploid HG002 genome to serve as a replacement for current variant-based benchmarks. More info here: github.com/marbl/HG002
31.10.2023 22:09 — 👍 31 🔁 22 💬 1 📌 1

If you discovered a novel microbial strain or species, please check out abon - a new program in the zol suite, which lets you easily check the conservation of your sample's BGC-ome (from antiSMASH & GECCO predictions) across target genomes: github.com/Kalan-Lab/zo... #secmet #bgc
27.10.2023 18:37 — 👍 13 🔁 15 💬 0 📌 0
Hope it will be helpful! We've tried SURVIVOR, which mostly worked good, but sometimes variants "disappear". Also there is Jasmin and truvari merge, but those were designed mostly for indels (e.g. germline SVs). Also rtg-tools has functinoality to compare BNDs.
25.10.2023 18:56 — 👍 2 🔁 0 💬 0 📌 0
Exactly! We actually did develop our own comparison method (for better or worse) - Asher is now tidying up the code and we'll make it available soon. It decomposes SVs from different tools into paired breakpoints and is agnostic to SV types. But overall, parsing different VCFs has been a pain..
24.10.2023 18:16 — 👍 3 🔁 0 💬 1 📌 0
Oups! We'll fix that!
24.10.2023 18:13 — 👍 2 🔁 0 💬 1 📌 0
Associate Professor
DFCI & HMS
Assistant professor @TGen | Postdoc @jacksonlab @MDAndersonNews | Genomics | Cancer | Gliomas | Telomeres
Mobilizing the fight for science and democracy, because Science is for everyone 🧪🌎
The hub for science activism!
Learn more ⬇️
http://linktr.ee/standupforscience
Bloomberg Distinguished Professor of BME, CS, and Biostats at Johns Hopkins Univ., tennis player, @StevenSalzberg1 on Twitter, lab: salzberg-lab.org, Substack blog: stevensalzberg.substack.com
Husband, Father and grandfather, Datahound, Dog lover, Fan of Celtic music, Former NIGMS director, Former EiC of Science magazine, Stand Up for Science advisor, Pittsburgh, PA
NIH Dashboard: https://jeremymberg.github.io/jeremyberg.github.io/index.html
Facts & strategy, in an authoritarian takeover.
Rightwing billionaires want to privatize NIH and use it to control universities.
We work to cure diseases like cancer.
Pers views. #science #medicine
Publishing the latest advances across all areas of cancer research and oncology. Part of @natureportfolio.nature.com
🌐 www.nature.com/natcancer/
📍New York, London, Berlin and Heidelberg
Original and probably disagreeable perspectives on genomics. Not sure when I'm joking and when I'm being serious. Opinions my own, not the herd’s.
Assistant Professor of Cancer Biology at Weill Cornell Medicine — NIH NCI R00 Funded — Chromatin & Epigenetics in Cancer and Immunity.
Group leader of the Stochastic Systems Biology Lab at IGBMC - CNRS - University of Strasbourg. Models of gene regulation based on biophysics-informed deep learning: https://www.igbmc.fr/molina
🔬 Cell adhesion networks lab
🥼 A new cell systems biology team
📍 University of Manchester, UK
🔗 https://linktr.ee/TheByronLab
https://www.helmholtz-munich.de/en/ife
Assistant Professor, Tish Cancer Institute @Mount Sinai. Colorectal Cancer. Oncofetal reprogramming. Views are my own. "By land and by sea, Carthage will rise again"
https://profiles.mountsinai.org/slim-mzoughi
Researcher in Epigenetics, CRISPRing around. MSCA Postdoctoral Fellow at EMBL Rome (Hackett lab). Before: Max Planck for Molecular Genetics (Ph.D.), Exeter Uni, Pasteur Institute, Sapienza Uni - Pessimismo dell' intelligenza/Ottimismo della volontà.
Lorry Lokey Chair & Professor of Bioengineering, Knight Campus, University of Oregon. evolution | ecology | genetics | genomics | evodevo | systems biology | epigenetics | host-microbe. Educational empowerment & increasing opportunities for all in science.
Post doc 🧬👩🏻🔬👩🏻💻 in @thecesarelab.bsky.social at @cmri.bsky.social | using insights from epigenetics and genome stability to improve pediatric cancer treatment | Optimist 😊 | Runner 🏃🏻♀️ | Wife & Mum 💛🌻
Developmental biologist working at CNRS/IGMM/University of Montpellier, interested by gene regulation in drosophila embryos. we use quantitative imaging to monitor mRNA lifecycle.
Group Leader at MPI Molgen, Berlin, Germany
Interested in quantitative biology, gene regulation, epigenetics, system biology and X-chromosome inactivation.
https://www.molgen.mpg.de/schulz-lab